Un-tagging proteins – it could make a world of difference!
Read the full article on ScienceDirect.
Regulation of human norovirus VPg nucleotidylylation by ProPol and nucleoside triphosphate binding by its amino terminal sequence in vitro
Text by Alexei Medvedev
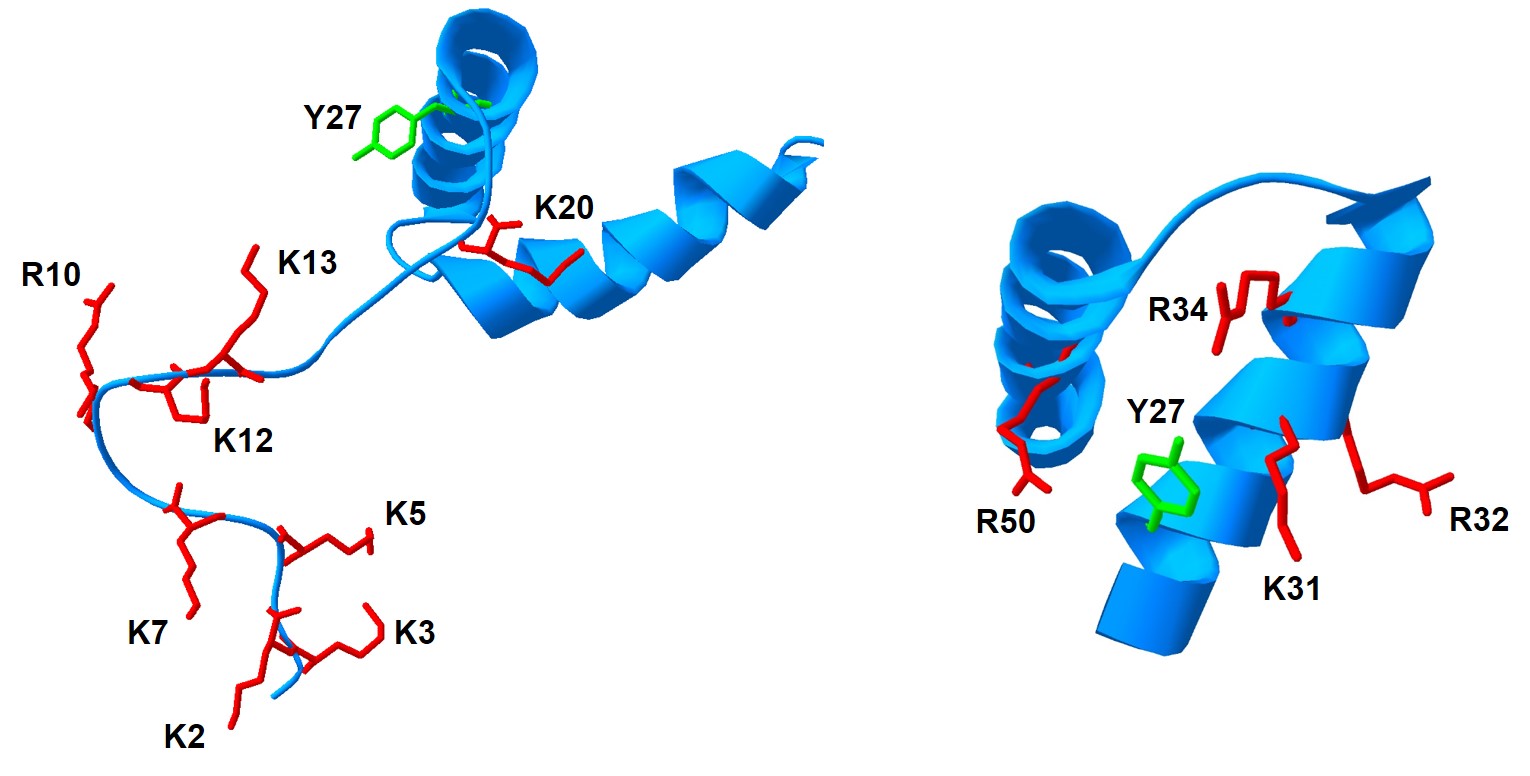
Human Norovirus (hNoV), the major cause of gastroenteritis outbreaks in the United States, is a single-stranded positive-sense RNA virus of Caliciviridae family. VPg is a multifunction protein covalently linked to 5’ end of the hNoV genome. The purpose of our study was to characterize the hNoV nucleotidylylation reaction that results in covalent attachment of nucleoside monophosphates to Tyr27 of VPg, a prerequisite to transcription and translation of the hNoV genome. Of particular interest was the ability of VPg to bind nucleotides and the influence positively charged amino acids in the N-terminus of VPg exert on nucleotidylylation. Over the course of the project, we demonstrated that ProPol is likely the primary polymerase responsible for hNoV VPg nucleotidylylation, that charged amino acids in the amino terminus of VPg regulate its nucleotidylylation, that nucleotides can be directly bound by hNoV VPg, and that VPg nucleotidylylation is likely regulated through several mechanisms.
Our study was partially based on work previously conducted on VPg nucleotidylylation of Potato Virus A, a Potyvirus, in which a lysine-rich nucleotide-binding site was discovered and shown to be essential for VPg nucleotidylylation, similar to our findings. Omission of a His6 tag from the N-terminus of the hNoV VPg in favor of a tag-less protein purification with the IMPACT kit (NEB) generated surprising results. VPg nucleotidylylation reaction was 70 times more effective in the absence of the His6 tag, and polymerase (Pol) precursor ProPol was 100 times superior to Pol at VPg nucleotidylylation in vitro, strongly implying that ProPol is likely the primary enzyme involved in hNoV VPg nucleotidylylation.
The most challenging part of our study was generating the 15 mutant hNoV VPg proteins required to complete the project. Each substitution was introduced using the Q5 Site-Directed Mutagenesis Kit (NEB). Constructs with multiple consecutive substitutions were especially challenging and time consuming to generate, as each round of point mutations was followed by cloning and sequencing before the next substitution could be introduced. Additionally, majority of the mutant proteins were expressed and purified in triplicate to ensure that experimental results were not a function of protein misfolding or other factors that could affect protein expression and purification. Optimization of tag-less VPg production and generation of VPg mutant library took about a year – were extra funds available, the mutant constructs could have been synthesized as IDT gBlocks and cloned into expression vectors for protein preparation in a fraction of that time.