Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation
Abstract
To expand the quantitative, systems level understanding and foster the expansion of the biotechnological applications of the filamentous bacteriophage M13, we have unified the accumulated quantitative information on M13 biology into a genetically-structured, experimentally-based computational simulation of the entire phage life cycle. The deterministic chemical kinetic simulation explicitly includes the molecular details of DNA replication, mRNA transcription, protein translation and particle assembly, as well as the competing protein-protein and protein-nucleic acid interactions that control the timing and extent of phage production. The simulation reproduces the holistic behavior of M13, closely matching experimentally reported values of the intracellular levels of phage species and the timing of events in the M13 life cycle. The computational model provides a quantitative description of phage biology, highlights gaps in the present understanding of M13, and offers a framework for exploring alternative mechanisms of regulation in the context of the complete M13 life cycle.
1. Introduction
Bacteriophages have been crucial to the development of molecular biology, serving as model systems for elucidating the central molecular mechanisms of living systems: DNA replication, transcription and translation. Increasingly, phage systems have been employed in biotechnological applications, most prominently in the identification and maturation of medically-relevant binding molecules through phage display (Ekiert et al., 2011, Ekiert et al., 2012, Karauzum et al., 2012, Koide and Sidhu, 2009, Smith, 1985). Phage display involves the creation and screening of large libraries of peptides or proteins displayed on the surface of the phage particle as a fusion to one of the coat proteins. Phage display libraries are passed over a target; particles that bind are retained. Because phages are biologically self-replicating systems, the particles that bind to the target may be amplified. Successive rounds of selection allow the evolution of tight binding molecules (Kay et al., 1996).The ability to select binding proteins is also central to many other applications of phages in materials and nanotechnology. The nanoscale size, simple life cycles, and speed and ease with which they can be prepared, manipulated and characterized render phages attractive and versatile biotechnological systems. Initial demonstrations that genetically-modified phage capsids could control the mineralization of metallic phases led to investigations of the ways in which biological molecules could be made to interact with and control the formation of inorganic materials and act as scaffolds for organizing chemical functionality on the nanoscale (Douglas and Young, 1998, Douglas and Young, 1999). Further developments have seen phage-based systems applied as scaffolds for tissue engineering (Wu et al., 2011, Yoo et al., 2011), as diagnostic imaging probes (Allen et al., 2005, Carrico et al., 2012, Ghosh et al., 2012), as drug delivery platforms (Flenniken et al., 2005, Rhee et al., 2012), and as replacements for antibodies in molecular diagnostics (Petrenko and Smith, 2000, Weiss and Penner, 2008). Phage systems remain at the forefront of research in nanomaterials and nanomedicine (Arter et al., 2010, Mao et al., 2004, Mao et al., 2011, Sanghvi et al., 2005, Steinmetz et al., 2011, Suthiwangcharoen et al., 2011, Udit et al., 2008, Yoo et al., 2006, Yoo et al., 2011).The ability to tune the properties of phage particles is critical to all of the uses of phages in nanomaterials and engineering. The filamentous bacteriophage of Escherichia coli (E. coli) are the most commonly employed system for phage display and are widely utilized in many biotechnological applications. The Ff filamentous phages f1, fd, and M13 are independent isolates of the same F-pilus specific phage. Kinetic parameters utilized in our simulation are derived from the combined experiments on f1, fd and M13. As M13 phage is most commonly used and our primary isolate of interest, we will use M13 throughout the manuscript. However, the simulation of the “M13” life cycle could be applied to any of the Ff phages. One of the key challenges for continued progress in developing highly modified M13 variants is the natural organization of the M13 genome. The M13 genome encodes 11 proteins with multiple overlapping genes, promoters, ribosome binding sites and terminators, and a complex control architecture that regulates the progression of infection (Fig. 1) (Cashman et al., 1980, Goodrich and Steege, 1999, Yen and Webster, 1982). Because of this complex organization, the independent manipulation of phage proteins leading to the controlled expression of fusion proteins on the surface of M13 particles can be difficult (Model and Russel, 1988, Rakonjac et al., 2011, Vanwezenbeek et al., 1980).
Engineering biology requires a detailed quantitative description of the molecular and systems level interactions governing at least transcription, translation, mRNA degradation, protein-protein, and protein-nucleic acid interactions. Data with this level of detail are woefully incomplete for most biological systems, but nearly complete for simple systems that have been intensively studied, such as M13 bacteriophage. Improvements in the quantitative understanding of biochemical systems are increasingly being leveraged to rationally expand biotechnological capabilities through synthetic biology. Synthetic biology is an approach to streamline and manage the complexity of biological systems through the application of an engineering design cycle to biology (Endy, 2005). A design cycle combines a detailed quantitative description of the system (allowing predictive design) with the ability to construct and evaluate designed variants. Knowledge gained and materials produced in each iteration of the cycle allow refinement of the model and leave a legacy infrastructure for the synthesis of improved variants. Phage systems are ideal targets for expanded development through synthetic biology (Baker et al., 2006, Endy, 2005, Voigt, 2012).This report describes the unification of the vast body of accumulated knowledge of filamentous phages into a genetically-structured, experimentally-based computational simulation of the life cycle of the non-lytic bacteriophage M13. We are motivated in developing a detailed computational model of the M13 life cycle to both better understand the complex biology of the phage and to serve as an aid in the design of phage systems with more rational control elements that will be more amenable to engineering.
1.1. Computational models of bacteriophages MS2, Qβ and T7
Models of the biology of phages and viruses typically address some of the aspects of the overall life cycle, such as the regulatory mechanisms in HIV (Hammond, 1993, Palsson et al., 1990) and lambda (Arkin et al., 1998, McAdams and Shapiro, 1995), the packaging of DNA in a phage capsid (Zlotnick, 1994), or the injection of DNA into a cell (Kindt et al., 2001, Tzlil et al., 2003). Several course grained models of filamentous phage infection have focused on horizontal gene transfer and the mechanism of conjugation inhibition by phage particles as well as the effects of cell growth on the heterogeneity of phage production (De Paepe et al., 2010, Lin et al., 2011, Wan et al., 2011, Wan and Goddard, 2012). More thorough life cycle models describing the complete process from infection to the release of progeny have been developed for the simple RNA phages MS2 and Qβ, whose genomes encode only 4 genes (Eigen et al., 1991, Eigen and Schuster, 1977, Kim and Yin, 2004, Tsukada et al., 2009).Complete models have also been developed for the more complex lytic bacteriophage T7 which contains 56 genes. Early models of T7 considered the connections between particular subsystems of DNA entry or gene expression (Buchholtz and Schneider, 1987, Maslak et al., 1993). The Yin laboratory improved on the early models to include all of the available information on the annotated functions of the T7 genome to build a simulation of the T7 life cycle (Endy et al., 1997, Endy and Yin, 2000, Endy et al., 2000, You and Yin, 2006, You et al., 2002, You and Yin, 1999, You and Yin, 2002). The initial T7 simulation combined general kinetic parameters involving host cellular processes such as DNA replication, RNA transcription and protein production with phage-specific processes such as the action of T7 RNA polymerases, functions of T7 proteins, and the assembly of progeny phages (Endy et al., 1997). Once developed, the T7 model was used to explore antiviral strategies that resist escape (Endy et al., 2000), to ask questions about the evolutionary fitness and systems level properties of the virus (Endy and Yin, 2000, You and Yin, 2000), and as the basis for exploring populations of phages in coupled infection/diffusion models (Duca et al., 2001, You and Yin, 1999). The model was adapted to a stochastic formulation to investigate the biases inherent in modeling discrete processes involving very small numbers of molecules as a continuous distribution (Srivastava et al., 2002). Finally, the model was used as a guide for refactoring T7 to separate overlapping genes and build a system more amenable to understanding and engineering (Chan et al., 2005).Inspired by the pioneering work of the Yin laboratory on the lytic phage T7, we sought to simulate the entire life cycle of the non-lytic phage M13. As with the T7 model, the M13 model is structured genetically to directly map the biological interactions to mathematical equations. Every parameter and interaction is a hypothesis about an aspect of the modeled system and is directly relatable to an experimentally measureable quantity. A genetically-structured model is distinct from unstructured phenomenological models which simply seek to find a mathematical form that fits some aspects of the available experimental data. Genetically-structured models are ideally suited to providing biological insight into complex systems, evaluating the consistency of the data used to produce them, and generating testable predictions about system behavior.The following sections describe the construction of a genetically-structured, in silico simulation of the M13 life cycle that combines experimental biochemical information generated over 50 years of study. The model was built from detailed descriptions of the M13 genome and the interactions of its components derived from many sources (Marvin and Hohn, 1969, Model and Russel, 1988, Rakonjac et al., 2011). The model describes the complete process of M13 replication from the initial entry of the phage single-stranded DNA genome into the cell to the production of progeny phages. The model explicitly includes the molecular components of phage DNA replication, the transcription and translation of phage mRNAs, and the functions of host- and phage-encoded proteins in the regulation of these processes. The model was parameterized using kinetic constants derived from experiments whenever available. Assumptions for the allocation of cellular resources to phage production were made using gross estimates of the cellular burden of phage infection based on the effect of phage infection on cell growth (Model et al., 1982).
1.2. M13 genome and general life cycle
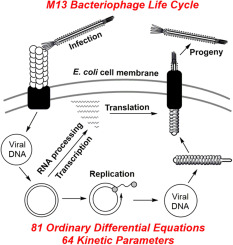
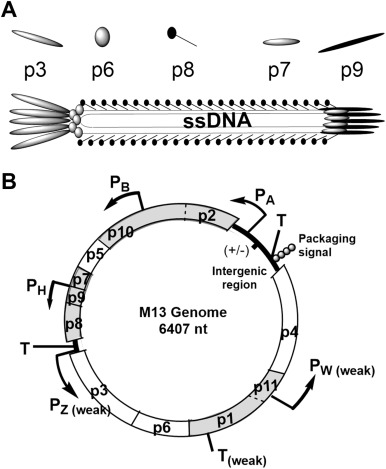
M13 is a filamentous phage that infects E. coli that carry the F-episome. Unlike the vast majority of bacterial viruses (e.g. MS2, Qβ, λ and T7), active infection with M13 does not kill the host cell, rather phage particles are continuously produced and extruded through the cell membrane as the infected cell continues to grow and divide (Young et al., 2000). The M13 phage particle consists of a single-stranded DNA (ssDNA) genome encased in approximately 2700 copies of a major coat protein, p8, and 5 copies each of 4 minor coat proteins: p3 and p6 on one end and p7 and p9 on the other (Fig. 1A). The M13 genes and corresponding proteins are named 1–11 and will be referred to as g1-g11 and p1–p11. The M13 genome is organized in two separate transcriptional units (Fig. 1B) (Vanwezenbeek et al., 1980). Together, five promoters and three terminators produce a cascade of mRNA species that contributes to the regulation of phage protein concentrations (Edens et al., 1978, Moses et al., 1980). An extended non-coding region in the M13 genome includes a promoter, a terminator, the origins of positive and negative strand DNA synthesis, as well as the signal that is required for packaging the ssDNA genome into a phage particle (Fig. 1B) (Dotto et al., 1981).The M13 life cycle (Fig. 2) begins with passage of the phage genome into a host cell in a poorly understood process mediated by coat protein p3 (Bennett et al., 2011). Upon entry into the cell, the ssDNA genome is rapidly converted to a double-stranded form by E. coli enzymes. The double-stranded replicative form DNA (RF DNA) immediately initiates mRNA transcription from the 5 constitutive promoters, producing mRNAs directing the synthesis of all 11 phage-encoded proteins. Additional single-stranded copies of the M13 genome are made through rolling circle replication using the RF DNA as a template. While concentrations of phage proteins are low, the single-stranded copies are converted into additional RF DNA. As the concentrations of phage proteins, particularly p5, increase, the rate of synthesis of RF DNA from ssDNA decreases. Instead, p5 binds to the ssDNA copies, preventing conversion into RF DNA and sequestering the ssDNA genomes for packaging into progeny phages. The p5-sequestered ssDNA is recognized by the membrane spanning phage assembly complex made up of phage proteins p1, an inner membrane spanning protein, p4, an outer membrane spanning protein, and p11, an inner membrane anchored periplasmic protein (Haigh and Webster, 1999, Marciano et al., 1999, Russel, 1993). Through a poorly understood process, the assembly complex first attaches the minor coat proteins p7 and p9 to the p5-sequestered ssDNA and then proceeds to pass the ssDNA through the cell membranes, concomitantly removing p5 and assembling p8 around the ssDNA genome. The process is completed by the addition of the other minor coat proteins p3 and p6, and the particle is released from the cell (Bennett et al., 2011).
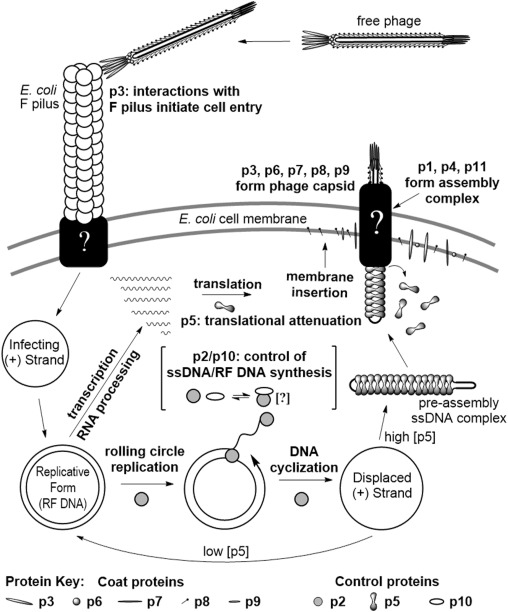
Fig. 2. Depiction of the M13 bacteriophage life cycle from cell entry to release of progeny phage. Question marks (?) indicate aspects of the life cycle for which the details of the molecular interactions are unknown.
2. Simulation
Many aspects of filamentous phage biology have been used as model systems to elucidate the molecular details of cellular phenomena, and these areas of intense study contributed significant quantitative data on M13 biology. A series of studies on the DNA replication, mRNA processing and mRNA degradation in M13 were particularly useful (Blumer et al., 1987, Blumer and Steege, 1982, Cashman et al., 1980, Goodrich and Steege, 1999, Meyer and Geider, 1982). The rates of typical E. coli cellular processes were collected from many sources; the T7 models and sources within (Endy et al., 1997) and bionumbers database (Milo et al., 2010) were of particular importance. In total 67% of the rate constants used in the model were estimated from experimental data (43 of 64 kinetic parameters in 81 differential equations. Model species are listed in Table 1, and the kinetic parameters used in the simulation are summarized in Table 2. The following sections describe the construction of the model with illustrative examples of the biochemical reactions (R#) and related kinetic equations. The kinetic equations in the model follow mass action kinetics. The differential equations that constitute the model were constructed by combining the relevant kinetic equations for each species across all of the reactions in which it is involved. Complete lists of the reactions, kinetic equations and differential equations are included in Supplementary Tables S1 and S2. The Matlab SimBiology file that contains the simulation is included as supplementary file; additional code that interfaces with Matlab to execute the simulation and perform automatic data manipulation is available upon request. Table S3 describes the major assumptions made when parameterizing the model. Table S4 highlights key areas of filamentous phage biology in which additional experiments would contribute substantively to an improved understanding of the phage life cycle.